NNX 和 NumPyro 集成
在此示例 notebook 中,我们将展示如何将来自 NNX 库的神经网络组件集成到 NumPyro 模型中。以类似的方式,您也可以使用 Flax Linen API。
此 notebook 基于博客文章 Flax and NumPyro Toy Example。
准备 Notebook
[1]:
!pip install -q numpyro@git+https://github.com/pyro-ppl/numpyro arviz flax matplotlib
[2]:
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
from flax import nnx
from jax import random
import jax.numpy as jnp
import numpyro
from numpyro.contrib.module import nnx_module, random_nnx_module
import numpyro.distributions as dist
from numpyro.handlers import condition
from numpyro.infer import SVI, Trace_ELBO
from numpyro.infer.autoguide import AutoNormal
from numpyro.infer.util import Predictive
plt.style.use("bmh")
plt.rcParams["figure.figsize"] = [10, 6]
plt.rcParams["figure.dpi"] = 100
plt.rcParams["figure.facecolor"] = "white"
numpyro.set_host_device_count(n=4)
rng_key = random.PRNGKey(seed=42)
%load_ext autoreload
%autoreload 2
%config InlineBackend.figure_format = "retina"
生成数据
我们生成了一个均值和标准差均非线性的合成数据集。
[3]:
n = 32 * 10
rng_key, rng_subkey = random.split(rng_key)
x = jnp.linspace(1, jnp.pi, n)
mu_true = jnp.sqrt(x + 0.5) * jnp.sin(9 * x)
sigma_true = 0.15 * x**2
rng_key, rng_subkey = random.split(rng_key)
y = mu_true + sigma_true * random.normal(rng_key, shape=(n,))
让我们可视化生成的数据集
[4]:
fig, ax = plt.subplots()
ax.plot(x, mu_true, color="C0", label=r"$\mu$", linewidth=3)
ax.fill_between(
x,
(mu_true - 2 * sigma_true),
(mu_true + 2 * sigma_true),
color="C0",
alpha=0.2,
label=r"$\mu \pm 2 \sigma$",
)
ax.scatter(x, y, color="black", label="data")
ax.legend(loc="upper left")
ax.set_title(label="Simulated Data", fontsize=18, fontweight="bold")
ax.set(xlabel="x", ylabel="y");
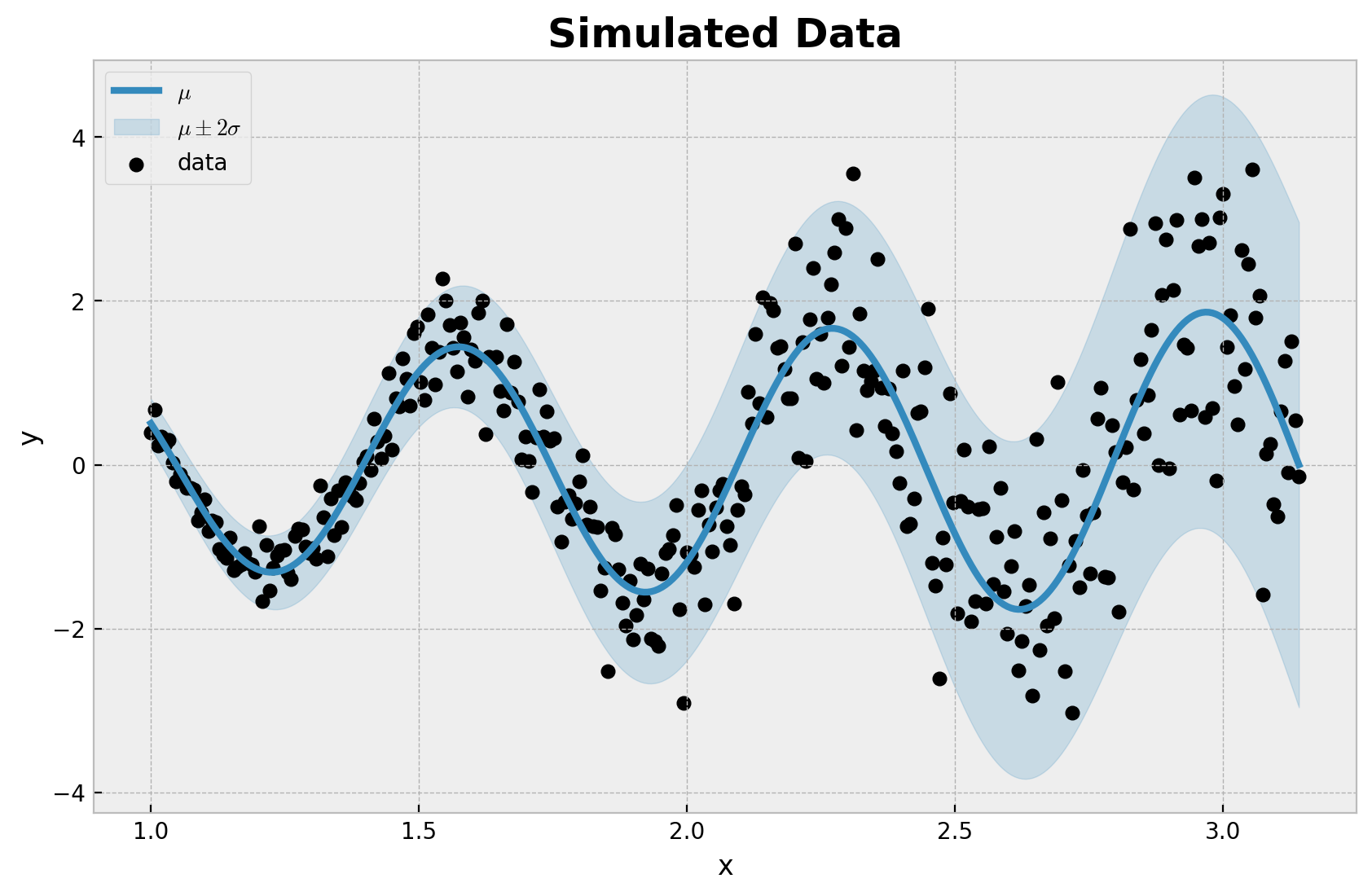
我们清楚地看到数据是非线性的,并且存在异方差噪声。我们希望使用神经网络对非线性进行建模,将数据的均值和标准差建模为输入 \(x\) 的函数。
模型规范
首先,我们准备训练数据。
[5]:
x_train = x[..., None]
y_train = y
接下来,我们使用 NNX 定义两个 MLP 组件,一个用于均值,一个用于标准差。您可以查看 `NNX basics <https://flax.jax.net.cn/en/v0.8.3/experimental/nnx/nnx_basics.html>`__ 以获取更多详细信息。
[6]:
class LocMLP(nnx.Module):
"""3-layer Multi-layer perceptron for the mean."""
def __init__(self, din: int, dmid: int, dout: int, *, rngs: nnx.Rngs):
self.linear1 = nnx.Linear(din, dmid, rngs=rngs)
self.linear2 = nnx.Linear(dmid, dmid, rngs=rngs)
self.linear3 = nnx.Linear(dmid, dout, rngs=rngs)
def __call__(self, x, rngs=None):
x = self.linear1(x)
x = nnx.sigmoid(x)
x = self.linear2(x)
x = nnx.sigmoid(x)
x = self.linear3(x)
return x
class ScaleMLP(nnx.Module):
"""Single-layer MLP for the standard deviation."""
def __init__(self, *, rngs: nnx.Rngs) -> None:
self.linear = nnx.Linear(1, 1, rngs=rngs)
def __call__(self, x, rngs=None):
x = self.linear(x)
return nnx.softplus(x)
现在我们以“即时”(eager)方式定义神经网络组件。
[7]:
mu_nn_module = LocMLP(din=1, dmid=8, dout=1, rngs=nnx.Rngs(0))
sigma_nn_module = ScaleMLP(rngs=nnx.Rngs(1))
最后,我们可以将神经网络组件添加到 NumPyro 模型中,我们在其中使用正态分布作为似然函数,并允许参数随输入 \(x\) 变化。
[8]:
def model(x):
# Neural network component for the mean. Here we consider the parameters of the
# neural network as learnable.
mu_nn = nnx_module("mu_nn", mu_nn_module)
# Here we consider the parameters of the neural network as random variables.
# Hence we can set priors for them.
sigma_nn = random_nnx_module(
"sigma_nn",
sigma_nn_module,
prior={
# From the data we know the variance is increasing over x.
# Hence we use a HalfNormal distribution to model the kernel term.
"linear.kernel": dist.HalfNormal(scale=1),
# We use a Normal distribution for the bias.
"linear.bias": dist.Normal(loc=0, scale=1),
},
)
mu = numpyro.deterministic("mu", mu_nn(x).squeeze())
sigma = numpyro.deterministic("sigma", sigma_nn(x).squeeze())
with numpyro.plate("data", len(x)):
numpyro.sample("likelihood", dist.Normal(loc=mu, scale=sigma))
numpyro.render_model(
model=model,
model_args=(x_train,),
render_distributions=True,
render_params=True,
)
[8]:
先验预测检查
在进行推断之前,我们可以检查先验预测分布,以确保先验是合理的。
[9]:
prior_predictive = Predictive(model=model, num_samples=100)
rng_key, rng_subkey = random.split(key=rng_key)
prior_predictive_samples = prior_predictive(rng_subkey, x_train)
obs_train = jnp.arange(x_train.size)
idata = az.from_dict(
prior_predictive={
k: np.expand_dims(a=np.asarray(v), axis=0)
for k, v in prior_predictive_samples.items()
},
coords={"obs": obs_train},
dims={"mu": ["obs"], "sigma": ["obs"], "likelihood": ["obs"]},
)
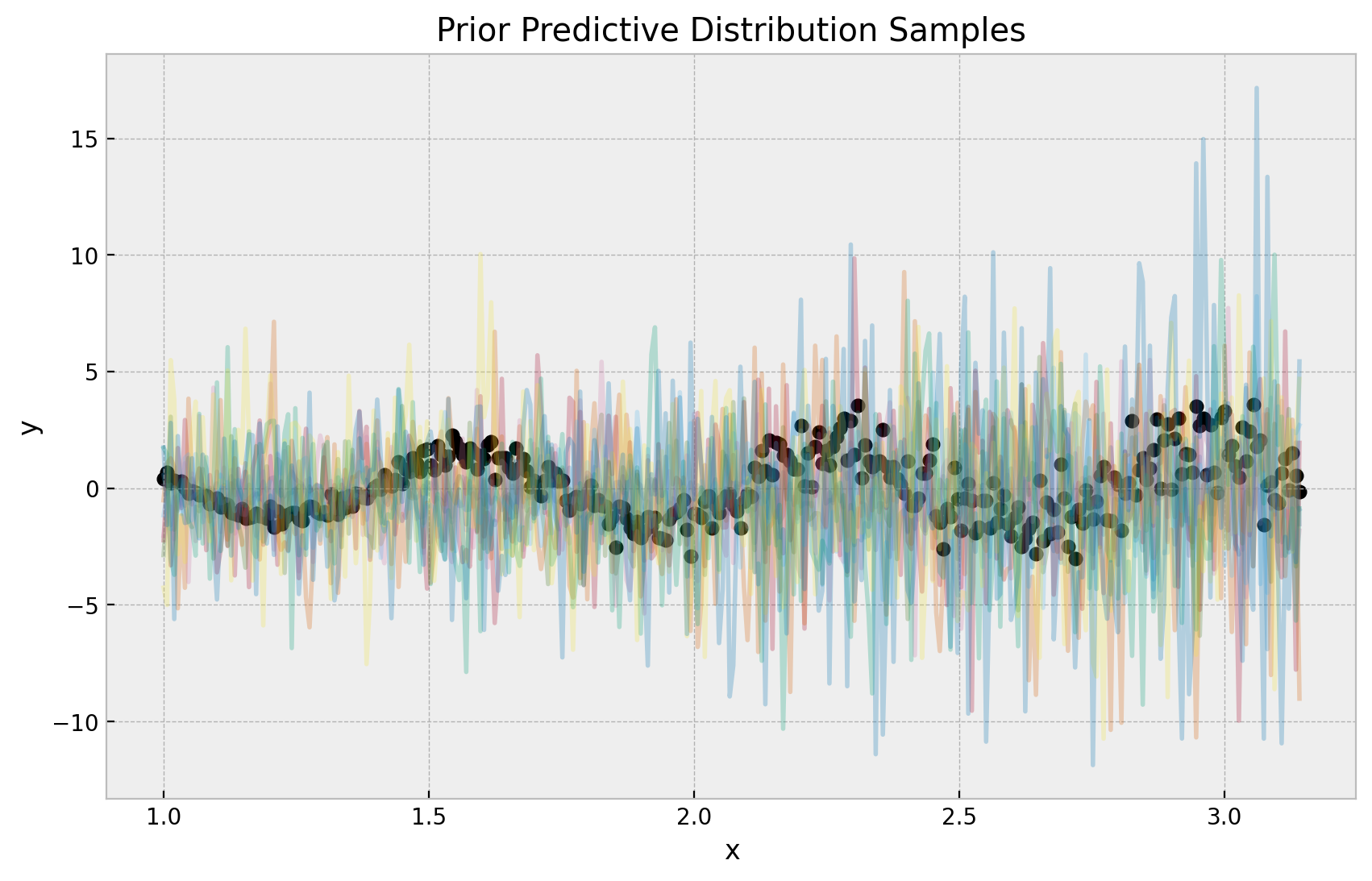
首先,我们可视化先验预测分布。我们清楚地看到方差随着 \(x\) 的增加而增加,正如我们预期的那样(由先验定义)。我们也看到样本的范围与数据在一个合理的范围内。
[10]:
fig, ax = plt.subplots()
for i in range(10):
ax.plot(x, idata["prior_predictive"]["likelihood"].sel(chain=0, draw=i), alpha=0.25)
ax.scatter(x, y, color="black", label="data")
ax.set(xlabel="x", ylabel="y", title="Prior Predictive Distribution Samples");
我们还可以验证 sigma 核的先验是否按预期工作
[11]:
fig, ax = plt.subplots()
az.plot_dist(
idata["prior_predictive"]["sigma_nn/linear.kernel"].squeeze(axis=(-1, -2)),
color="C2",
fill_kwargs={"alpha": 0.3},
)
ax.set(
xlabel="sigma_nn/linear.kernel",
title="Prior Predictive Distribution - Kernel of the Standard Deviation MLP",
);
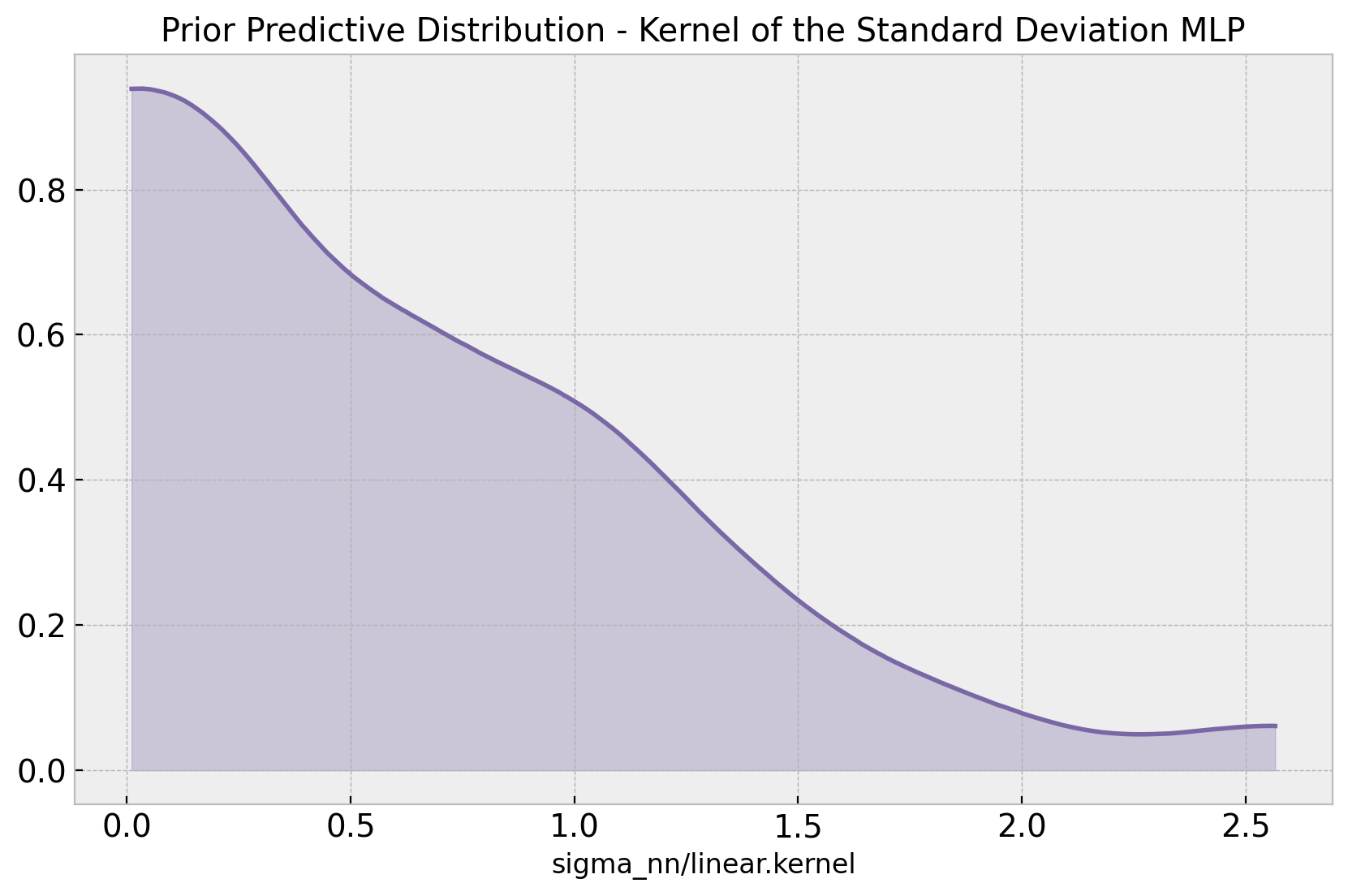
我们确实看到先验是正的!
模型推断
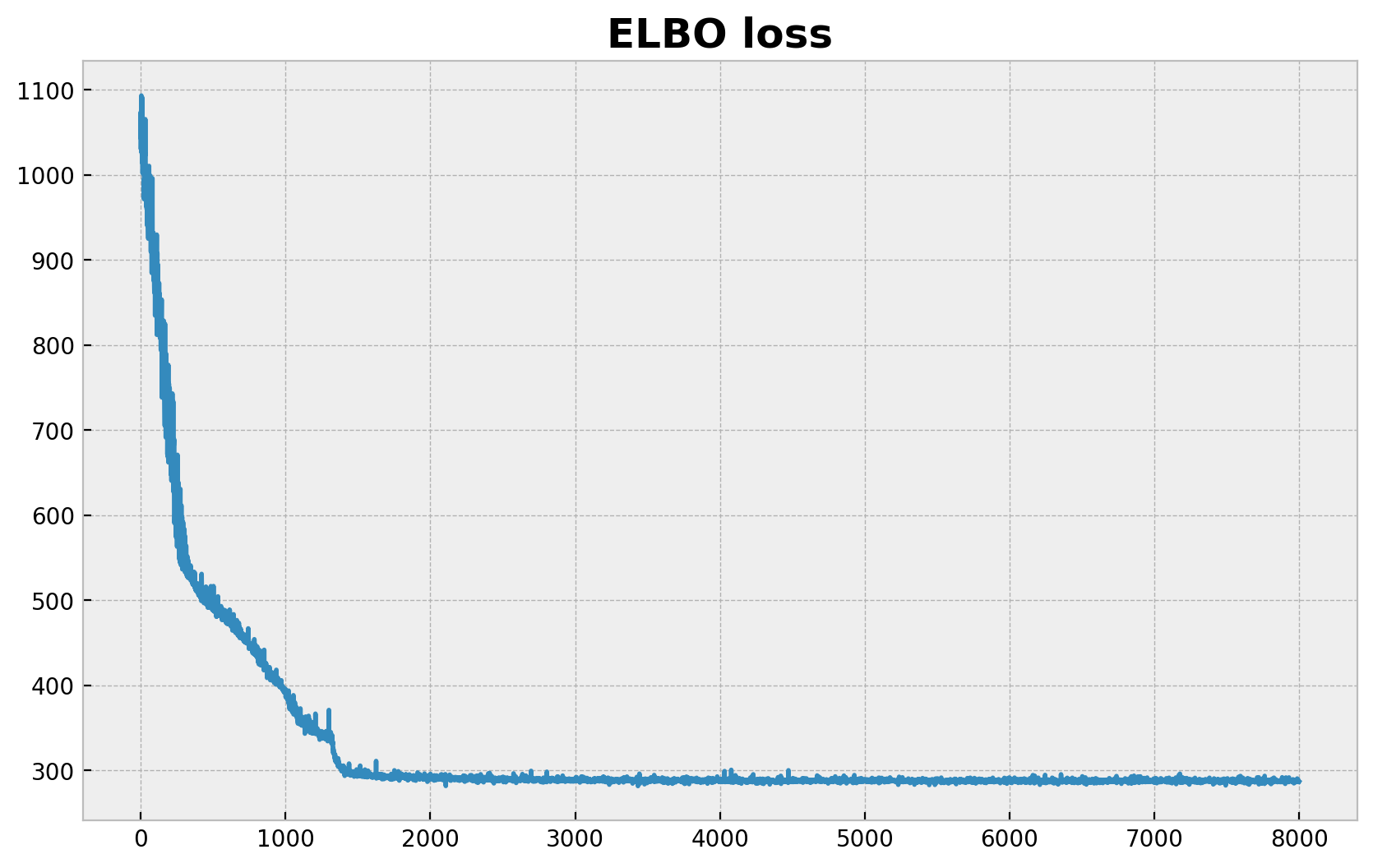
现在我们使用 SVI 对模型进行推断。
[12]:
# We condition the model on the training data
conditioned_model = condition(model, data={"likelihood": y_train})
guide = AutoNormal(model=conditioned_model)
optimizer = numpyro.optim.Adam(step_size=0.005)
svi = SVI(conditioned_model, guide, optimizer, loss=Trace_ELBO())
n_samples = 8_000
rng_key, rng_subkey = random.split(key=rng_key)
svi_result = svi.run(rng_subkey, n_samples, x_train)
fig, ax = plt.subplots()
ax.plot(svi_result.losses)
ax.set_title("ELBO loss", fontsize=18, fontweight="bold");
100%|██████████| 8000/8000 [00:01<00:00, 5976.75it/s, init loss: 1032.5969, avg. loss [7601-8000]: 287.5595]
现在我们生成后验预测分布。
[13]:
params = svi_result.params
posterior_predictive = Predictive(
model=model,
guide=guide,
params=params,
num_samples=2_000,
return_sites=["mu", "sigma", "likelihood"],
)
rng_key, rng_subkey = random.split(key=rng_key)
posterior_predictive_samples = posterior_predictive(rng_subkey, x_train)
现在我们收集后验预测样本以便进行可视化。
[14]:
idata.extend(
az.from_dict(
posterior_predictive={
k: np.expand_dims(a=np.asarray(v), axis=0)
for k, v in posterior_predictive_samples.items()
},
coords={"obs": obs_train},
dims={"mu": ["obs"], "sigma": ["obs"], "likelihood": ["obs"]},
)
)
最后,我们可视化后验预测分布、均值和标准差组件。
[15]:
fig, ax = plt.subplots(
nrows=3,
ncols=1,
sharex=True,
sharey=True,
figsize=(10, 9),
layout="constrained",
)
az.plot_hdi(
x,
idata["posterior_predictive"]["likelihood"],
color="C1",
fill_kwargs={"alpha": 0.3, "label": "94% HDI"},
ax=ax[0],
)
ax[0].plot(
x_train,
idata["posterior_predictive"]["likelihood"].mean(dim=("chain", "draw")),
color="C1",
linewidth=3,
label="SVI Posterior Mean",
)
ax[0].plot(x, mu_true, color="C0", label=r"$\mu$", linewidth=3)
ax[0].scatter(x, y, color="black", label="data")
ax[0].legend(loc="upper left")
ax[0].set(ylabel="y")
ax[0].set_title(label="Posterior Predictive Distribution")
ax[1].plot(
x,
idata["posterior_predictive"]["mu"].mean(dim=("chain", "draw")),
linewidth=3,
color="C2",
)
ax[1].set_title(label="Mean")
ax[2].plot(
x,
idata["posterior_predictive"]["sigma"].mean(dim=("chain", "draw")),
linewidth=3,
color="C3",
)
ax[2].set(xlabel="x")
ax[2].set_title(label="Standard Deviation");
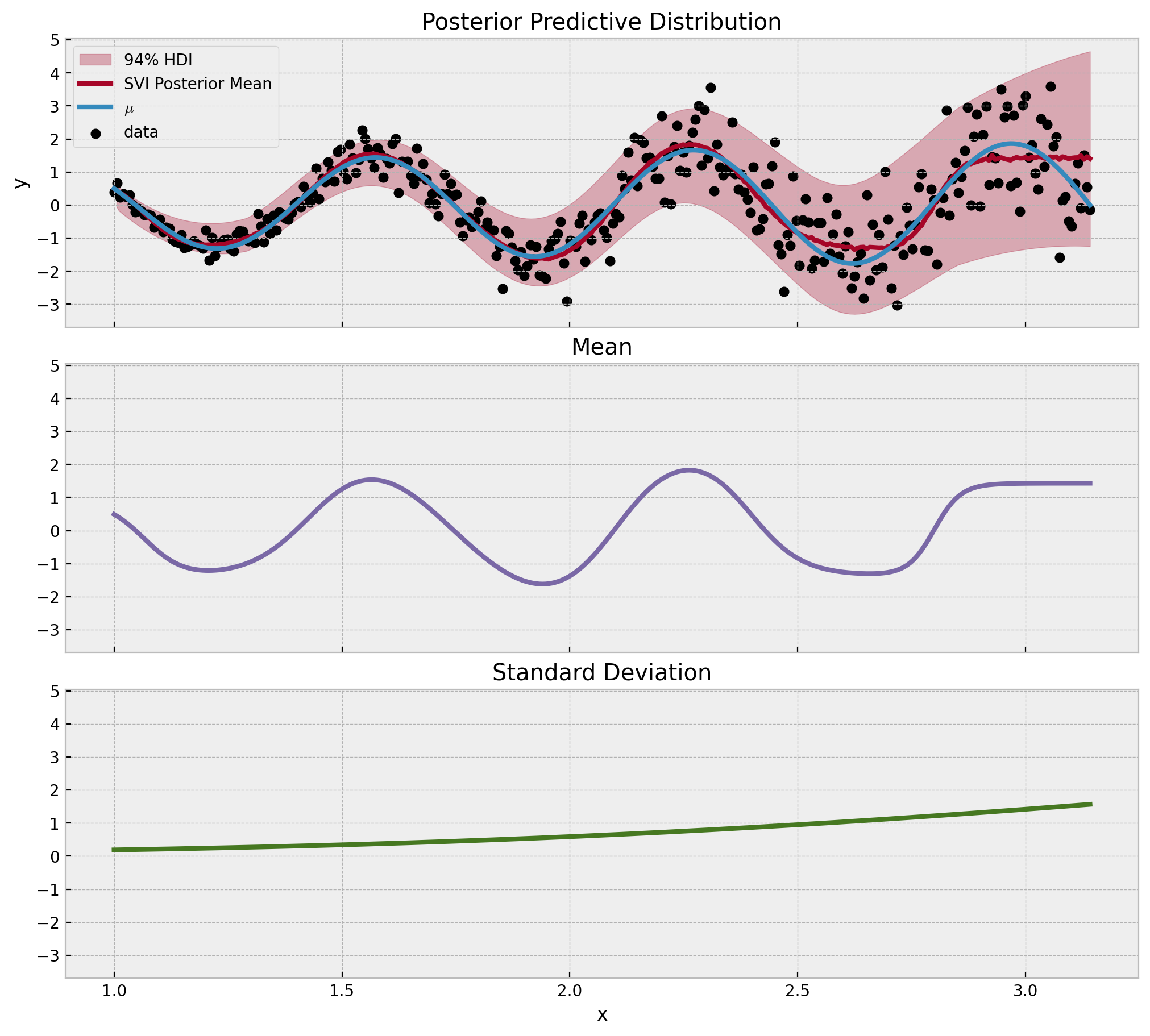
结果看起来很棒!拟合和组件都按预期工作。