注意
转到末尾 下载完整示例代码。
示例:神经传输
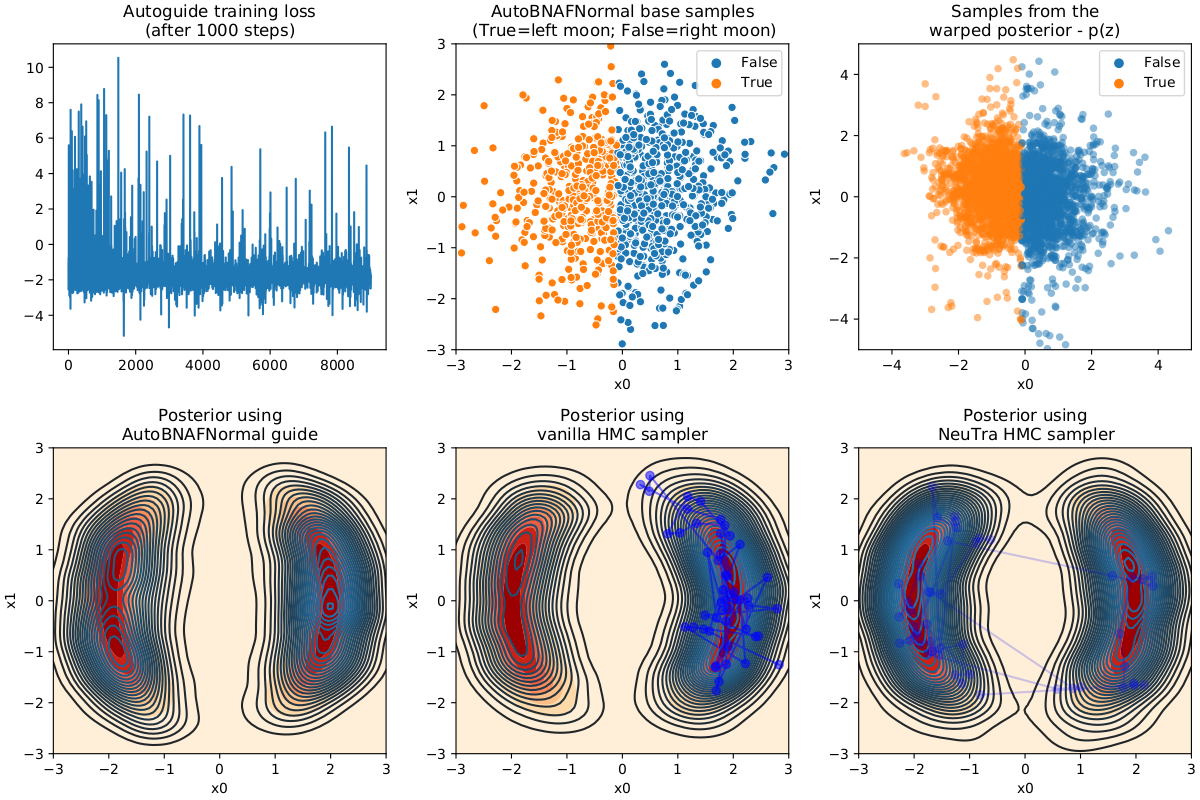
本示例演示如何使用训练好的 AutoBNAFNormal autoguide 将后验分布转换为类似高斯分布。此变换将用于改善 NUTS 采样器的混合效率。
参考文献
Hoffman, M. et al. (2019), “NeuTra-lizing Bad Geometry in Hamiltonian Monte Carlo Using Neural Transport”, (https://arxiv.org/abs/1903.03704)
import argparse
import os
from matplotlib.gridspec import GridSpec
import matplotlib.pyplot as plt
import seaborn as sns
from jax import random
import jax.numpy as jnp
from jax.scipy.special import logsumexp
import numpyro
from numpyro import optim
from numpyro.diagnostics import print_summary
import numpyro.distributions as dist
from numpyro.distributions import constraints
from numpyro.infer import MCMC, NUTS, SVI, Trace_ELBO
from numpyro.infer.autoguide import AutoBNAFNormal
from numpyro.infer.reparam import NeuTraReparam
class DualMoonDistribution(dist.Distribution):
support = constraints.real_vector
def __init__(self):
super(DualMoonDistribution, self).__init__(event_shape=(2,))
def sample(self, key, sample_shape=()):
# it is enough to return an arbitrary sample with correct shape
return jnp.zeros(sample_shape + self.event_shape)
def log_prob(self, x):
term1 = 0.5 * ((jnp.linalg.norm(x, axis=-1) - 2) / 0.4) ** 2
term2 = -0.5 * ((x[..., :1] + jnp.array([-2.0, 2.0])) / 0.6) ** 2
pe = term1 - logsumexp(term2, axis=-1)
return -pe
def dual_moon_model():
numpyro.sample("x", DualMoonDistribution())
def main(args):
print("Start vanilla HMC...")
nuts_kernel = NUTS(dual_moon_model)
mcmc = MCMC(
nuts_kernel,
num_warmup=args.num_warmup,
num_samples=args.num_samples,
num_chains=args.num_chains,
progress_bar=False if "NUMPYRO_SPHINXBUILD" in os.environ else True,
)
mcmc.run(random.PRNGKey(0))
mcmc.print_summary()
vanilla_samples = mcmc.get_samples()["x"].copy()
guide = AutoBNAFNormal(
dual_moon_model, hidden_factors=[args.hidden_factor, args.hidden_factor]
)
svi = SVI(dual_moon_model, guide, optim.Adam(0.003), Trace_ELBO())
print("Start training guide...")
svi_result = svi.run(random.PRNGKey(1), args.num_iters)
print("Finish training guide. Extract samples...")
guide_samples = guide.sample_posterior(
random.PRNGKey(2), svi_result.params, sample_shape=(args.num_samples,)
)["x"].copy()
print("\nStart NeuTra HMC...")
neutra = NeuTraReparam(guide, svi_result.params)
neutra_model = neutra.reparam(dual_moon_model)
nuts_kernel = NUTS(neutra_model)
mcmc = MCMC(
nuts_kernel,
num_warmup=args.num_warmup,
num_samples=args.num_samples,
num_chains=args.num_chains,
progress_bar=False if "NUMPYRO_SPHINXBUILD" in os.environ else True,
)
mcmc.run(random.PRNGKey(3))
mcmc.print_summary()
zs = mcmc.get_samples(group_by_chain=True)["auto_shared_latent"]
print("Transform samples into unwarped space...")
samples = neutra.transform_sample(zs)
print_summary(samples)
zs = zs.reshape(-1, 2)
samples = samples["x"].reshape(-1, 2).copy()
# make plots
# guide samples (for plotting)
guide_base_samples = dist.Normal(jnp.zeros(2), 1.0).sample(
random.PRNGKey(4), (1000,)
)
guide_trans_samples = neutra.transform_sample(guide_base_samples)["x"]
x1 = jnp.linspace(-3, 3, 100)
x2 = jnp.linspace(-3, 3, 100)
X1, X2 = jnp.meshgrid(x1, x2)
P = jnp.exp(DualMoonDistribution().log_prob(jnp.stack([X1, X2], axis=-1)))
fig = plt.figure(figsize=(12, 8), constrained_layout=True)
gs = GridSpec(2, 3, figure=fig)
ax1 = fig.add_subplot(gs[0, 0])
ax2 = fig.add_subplot(gs[1, 0])
ax3 = fig.add_subplot(gs[0, 1])
ax4 = fig.add_subplot(gs[1, 1])
ax5 = fig.add_subplot(gs[0, 2])
ax6 = fig.add_subplot(gs[1, 2])
ax1.plot(svi_result.losses[1000:])
ax1.set_title("Autoguide training loss\n(after 1000 steps)")
ax2.contourf(X1, X2, P, cmap="OrRd")
sns.kdeplot(x=guide_samples[:, 0], y=guide_samples[:, 1], n_levels=30, ax=ax2)
ax2.set(
xlim=[-3, 3],
ylim=[-3, 3],
xlabel="x0",
ylabel="x1",
title="Posterior using\nAutoBNAFNormal guide",
)
sns.scatterplot(
x=guide_base_samples[:, 0],
y=guide_base_samples[:, 1],
ax=ax3,
hue=guide_trans_samples[:, 0] < 0.0,
)
ax3.set(
xlim=[-3, 3],
ylim=[-3, 3],
xlabel="x0",
ylabel="x1",
title="AutoBNAFNormal base samples\n(True=left moon; False=right moon)",
)
ax4.contourf(X1, X2, P, cmap="OrRd")
sns.kdeplot(x=vanilla_samples[:, 0], y=vanilla_samples[:, 1], n_levels=30, ax=ax4)
ax4.plot(vanilla_samples[-50:, 0], vanilla_samples[-50:, 1], "bo-", alpha=0.5)
ax4.set(
xlim=[-3, 3],
ylim=[-3, 3],
xlabel="x0",
ylabel="x1",
title="Posterior using\nvanilla HMC sampler",
)
sns.scatterplot(
x=zs[:, 0],
y=zs[:, 1],
ax=ax5,
hue=samples[:, 0] < 0.0,
s=30,
alpha=0.5,
edgecolor="none",
)
ax5.set(
xlim=[-5, 5],
ylim=[-5, 5],
xlabel="x0",
ylabel="x1",
title="Samples from the\nwarped posterior - p(z)",
)
ax6.contourf(X1, X2, P, cmap="OrRd")
sns.kdeplot(x=samples[:, 0], y=samples[:, 1], n_levels=30, ax=ax6)
ax6.plot(samples[-50:, 0], samples[-50:, 1], "bo-", alpha=0.2)
ax6.set(
xlim=[-3, 3],
ylim=[-3, 3],
xlabel="x0",
ylabel="x1",
title="Posterior using\nNeuTra HMC sampler",
)
plt.savefig("neutra.pdf")
if __name__ == "__main__":
assert numpyro.__version__.startswith("0.18.0")
parser = argparse.ArgumentParser(description="NeuTra HMC")
parser.add_argument("-n", "--num-samples", nargs="?", default=4000, type=int)
parser.add_argument("--num-warmup", nargs="?", default=1000, type=int)
parser.add_argument("--num-chains", nargs="?", default=1, type=int)
parser.add_argument("--hidden-factor", nargs="?", default=8, type=int)
parser.add_argument("--num-iters", nargs="?", default=10000, type=int)
parser.add_argument("--device", default="cpu", type=str, help='use "cpu" or "gpu".')
args = parser.parse_args()
numpyro.set_platform(args.device)
numpyro.set_host_device_count(args.num_chains)
main(args)