注意
跳转至末尾下载完整示例代码。
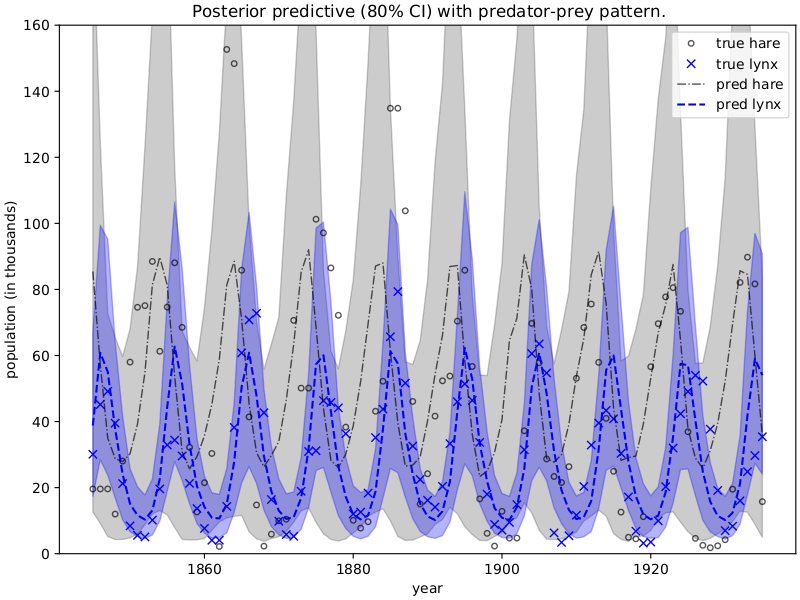
示例:捕食者-猎物模型
本示例复现了出色的案例研究 [1],该研究利用 Lotka-Volterra 方程 [2] 描述了加拿大猞猁(捕食者)和雪鞋兔(猎物)种群的动态。我们将使用从 [3] 获取的数据集,并运行 MCMC 来推断控制动态的微分方程的参数。
参考文献
import argparse
import os
import matplotlib
import matplotlib.pyplot as plt
from jax.experimental.ode import odeint
import jax.numpy as jnp
from jax.random import PRNGKey
import numpyro
import numpyro.distributions as dist
from numpyro.examples.datasets import LYNXHARE, load_dataset
from numpyro.infer import MCMC, NUTS, Predictive
matplotlib.use("Agg") # noqa: E402
def dz_dt(z, t, theta):
"""
Lotka–Volterra equations. Real positive parameters `alpha`, `beta`, `gamma`, `delta`
describes the interaction of two species.
"""
u = z[0]
v = z[1]
alpha, beta, gamma, delta = (
theta[..., 0],
theta[..., 1],
theta[..., 2],
theta[..., 3],
)
du_dt = (alpha - beta * v) * u
dv_dt = (-gamma + delta * u) * v
return jnp.stack([du_dt, dv_dt])
def model(N, y=None):
"""
:param int N: number of measurement times
:param numpy.ndarray y: measured populations with shape (N, 2)
"""
# initial population
z_init = numpyro.sample("z_init", dist.LogNormal(jnp.log(10), 1).expand([2]))
# measurement times
ts = jnp.arange(float(N))
# parameters alpha, beta, gamma, delta of dz_dt
theta = numpyro.sample(
"theta",
dist.TruncatedNormal(
low=0.0,
loc=jnp.array([1.0, 0.05, 1.0, 0.05]),
scale=jnp.array([0.5, 0.05, 0.5, 0.05]),
),
)
# integrate dz/dt, the result will have shape N x 2
z = odeint(dz_dt, z_init, ts, theta, rtol=1e-6, atol=1e-5, mxstep=1000)
# measurement errors
sigma = numpyro.sample("sigma", dist.LogNormal(-1, 1).expand([2]))
# measured populations
numpyro.sample("y", dist.LogNormal(jnp.log(z), sigma), obs=y)
def main(args):
_, fetch = load_dataset(LYNXHARE, shuffle=False)
year, data = fetch() # data is in hare -> lynx order
# use dense_mass for better mixing rate
mcmc = MCMC(
NUTS(model, dense_mass=True),
num_warmup=args.num_warmup,
num_samples=args.num_samples,
num_chains=args.num_chains,
progress_bar=False if "NUMPYRO_SPHINXBUILD" in os.environ else True,
)
mcmc.run(PRNGKey(1), N=data.shape[0], y=data)
mcmc.print_summary()
# predict populations
pop_pred = Predictive(model, mcmc.get_samples())(PRNGKey(2), data.shape[0])["y"]
mu = jnp.mean(pop_pred, 0)
pi = jnp.percentile(pop_pred, jnp.array([10, 90]), 0)
plt.figure(figsize=(8, 6), constrained_layout=True)
plt.plot(year, data[:, 0], "ko", mfc="none", ms=4, label="true hare", alpha=0.67)
plt.plot(year, data[:, 1], "bx", label="true lynx")
plt.plot(year, mu[:, 0], "k-.", label="pred hare", lw=1, alpha=0.67)
plt.plot(year, mu[:, 1], "b--", label="pred lynx")
plt.fill_between(year, pi[0, :, 0], pi[1, :, 0], color="k", alpha=0.2)
plt.fill_between(year, pi[0, :, 1], pi[1, :, 1], color="b", alpha=0.3)
plt.gca().set(ylim=(0, 160), xlabel="year", ylabel="population (in thousands)")
plt.title("Posterior predictive (80% CI) with predator-prey pattern.")
plt.legend()
plt.savefig("ode_plot.pdf")
if __name__ == "__main__":
assert numpyro.__version__.startswith("0.18.0")
parser = argparse.ArgumentParser(description="Predator-Prey Model")
parser.add_argument("-n", "--num-samples", nargs="?", default=1000, type=int)
parser.add_argument("--num-warmup", nargs="?", default=1000, type=int)
parser.add_argument("--num-chains", nargs="?", default=1, type=int)
parser.add_argument("--device", default="cpu", type=str, help='use "cpu" or "gpu".')
args = parser.parse_args()
numpyro.set_platform(args.device)
numpyro.set_host_device_count(args.num_chains)
main(args)